Deep learning has transformed medical image segmentation, yet its performance often declines when applied to data from unfamiliar domains. This is especially problematic in abdominal imaging, where variability across scanners, protocols and patient demographics can limit the reliability of automated tools. To address this challenge, a new model has been developed using domain randomisation in both image and feature spaces. Built on the nnU-Net framework, the model aims to improve the generalisability and accuracy of segmentation across CT and MRI scans. The method was validated on multiple datasets and benchmarked against leading tools, showing significant improvements, particularly in handling domain shifts.
Addressing Domain Shifts in Medical Imaging
Medical image segmentation models often fail when applied to data that differ from their training sets. This issue, known as domain shift, arises due to differences in imaging devices, acquisition settings and patient characteristics. In practice, such variation is the norm rather than the exception. Domain Generalisation (DG) seeks to counter this by training models that can generalise to unseen data. A promising approach within DG is domain randomisation, which introduces artificial variability during training. By simulating unseen conditions, models learn to focus on stable, domain-invariant features. In this context, high-level anatomical features are more consistent across domains than low-level image characteristics, such as texture or intensity. The new method builds on this insight by applying randomisation techniques to both the pixel-level intensity (image space) and internal network representations (feature space).
Must Read: Advancing Multiorgan Segmentation for Abdominal MRI
In the image space, the model modifies organ and tissue intensities using a Bézier curve-based transformation. This allows realistic yet diverse augmentation while preserving anatomical structure. A saliency-balancing mechanism ensures that the transformations remain clinically plausible. In the feature space, random perturbations are applied to low-level features via controlled changes in their statistical properties. By doing so, the model learns to rely less on unstable low-level textures and more on consistent anatomical features, improving generalisability across modalities and institutions.
Experimental Validation and Performance Assessment
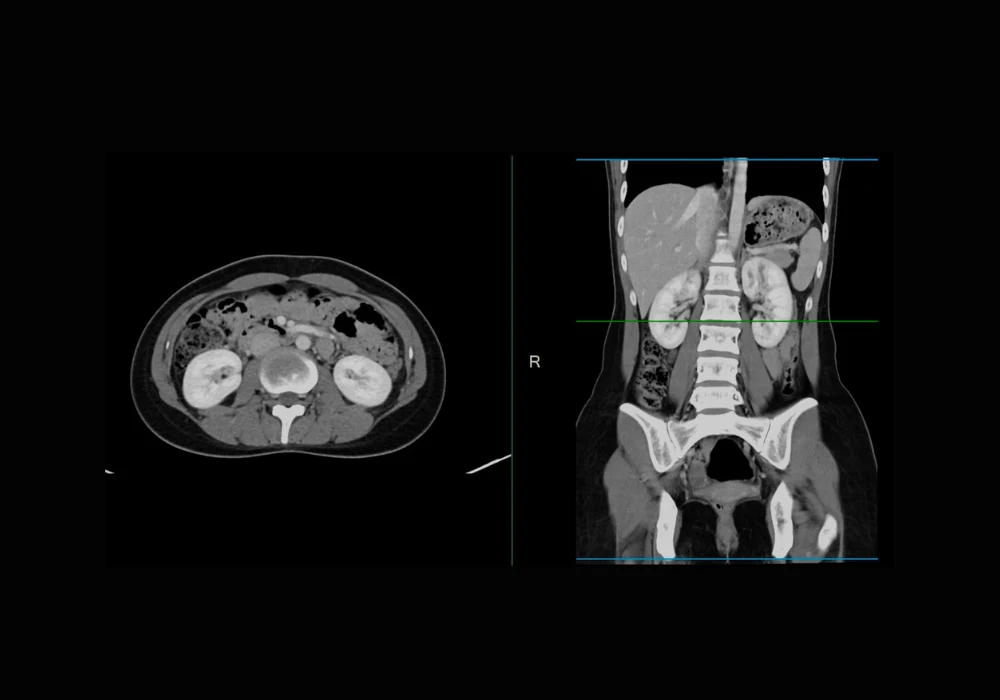
The model was tested on multiple public datasets under both cross-site and cross-modality conditions. Cross-site generalisation involved training the model on data from several institutions and evaluating it on previously unseen sites. Cross-modality testing involved training on one imaging modality (such as MRI) and testing on another (such as CT). The model was further assessed through ablation studies and comparisons with existing segmentation methods.
In cross-site experiments on prostate MRI datasets, the new model outperformed several established DG methods. Its performance, measured by the Dice Similarity Coefficient (DSC), consistently ranked highest across all tested domains. In cross-modality settings, the model achieved an average DSC of 0.84 when transferring from MRI to CT, surpassing the nearest competitor by 0.01. Particularly notable was its improvement in spleen segmentation, which showed a 0.04 gain in DSC. When transferring from CT to MRI, the model maintained parity with top-performing alternatives, highlighting its balanced generalisation capabilities.
An ablation study tested four configurations: no randomisation, image space only, feature space only and both spaces combined. Each component improved performance individually, but the combined strategy yielded the best results, reaching an average DSC of 0.88. These findings confirm that dual-space randomisation offers complementary benefits and is more effective than using either space alone.
Benchmarking Against Leading Segmentation Tools
The model’s performance was compared with TotalSegmentator and MRSegmentator, two widely used tools based on the same nnU-Net backbone but trained on large-scale datasets without domain randomisation. Despite the extensive training of these tools, the new model achieved higher segmentation accuracy on unseen test domains. In average DSC scores, it reached 0.88, compared to 0.76 for TotalSegmentator and 0.79 for MRSegmentator. The statistical significance of these differences was confirmed by ANOVA and posthoc analyses.
Performance gains were especially apparent for challenging organs such as the pancreas, adrenal glands and duodenum. These structures often pose difficulties due to low contrast or anatomical complexity. The new model delivered better boundary delineation, greater completeness and clearer separation of adjacent structures. Visual comparisons on 2D slices and 3D volumes further supported these quantitative findings. In terms of runtime and resource usage, all methods showed comparable efficiency, underscoring that the performance gains did not come at the cost of higher computational demand.
The study introduced a segmentation model that integrates domain randomisation in both image and feature spaces, significantly enhancing generalisability across CT and MRI data. Through rigorous validation, the method demonstrated consistent improvements over established tools and techniques. It achieved superior accuracy in both cross-site and cross-modality settings, particularly for anatomically complex organs.
By leveraging the strengths of both CT and MRI, as well as combining domain randomisation techniques, the model presents a robust and clinically adaptable solution. Its potential extends beyond abdominal imaging to other domains such as breast, musculoskeletal and neuroimaging. While limitations remain—such as the relative scarcity of diverse MRI datasets and the risks of over-randomisation—the results offer a promising step forward. With further refinement, this approach could facilitate more reliable and widespread adoption of AI-driven segmentation in varied clinical settings.
Source: Radiology: Artificial intelligence
Image Credit: iStock