HealthManagement, Volume 23 - Issue 4, 2023
Ongoing research and challenges regarding the use of Artificial Intelligence and Machine Learning to leverage data and identify complex interactions to address antimicrobial resistance in the ICU.
The possibilities of artificial intelligence (AI), and more specifically, machine learning (ML), are being researched across almost all domains of medicine, and the field of intensive care medicine is certainly no exception. Several factors contribute to the intensive care unit (ICU) being one of the focal points of AI/ML research. Intensive care was one of the earliest adaptors of the electronic health record (EHR) (Varun and Marik 2002), making data readily available. A vast amount of diverse data is generated for an individual patient during clinical care, resulting in 6.000–30.000 routinely collected healthcare data points per ICU patient per day. As this data is mostly structured and annotated, it is very suitable for training ML models. Although these datasets still need to be unbolted to be usable for (big) data research in many cases, several readyto-use publicly available datasets extracted from information in the EHR already exist to date (Sauer et al. 2022; Jin et al. 2023; Rodemund et al. 2023). At the same time, large nationwide data-sharing initiatives are being set up to unlock even more routinely collected healthcare data for clinicians, researchers and data scientists alike. As such, the data-rich ICU environment has and is developing the prerequisites to further enable (big) data research. The expectations for the clinical impact of AI have risen alongside the number of published studies. It is generally expected that AI will harbour benefits for patients as well as clinicians and the whole of society (Topol 2019). This is reflected in the variety of study aims for which AI has been investigated in published studies so far (van de Sande et al. 2021).
From a societal, patient, and research perspective, antimicrobial resistance (AMR) is an interesting research area for AI/ML, as it remains one of the biggest threats to global healthcare. According to the O'Neill report, it is estimated that by 2050, the death toll due to AMR could rise to 10 million lives a year worldwide (O'Neill 2023). AMR is particularly relevant for the ICU patient, as several AMR drivers (e.g., high selective pressure due to frequent antimicrobial use) are present in the ICU, and AMR infections can heavily impact a patient's ICU morbidity and mortality. One way of tackling the increase of AMR is by antimicrobial stewardship, which aims to combat antibiotic resistance by improving antimicrobial prescribing and use (CDC 2022). Antimicrobial stewardship comprises several interventions that can be performed during every step of antimicrobial therapy, and several use cases have been researched to explore how software and/or AI might aid in 'the rights' of antimicrobial prescription: the right drug at the right time and the right dose for the right patient (Grissinger 2010).
Identifying the right ICU patient means identifying the ICU patient that has a higher probability of infection than for inflammation. At present, clinicians use a combination of clinical gestalt and clinical decision rules when making this discrimination by incorporating information from the medical history, clinical examination, technical investigations, and laboratory and microbiological information. Research has shown that differentiating infection from inflammation poses a challenge for ICU physicians, especially when the infection is defined by consensus criteria, for instance, in the case of sepsis or ventilated-associated Ongoing research and challenges regarding the use of Artificial Intelligence and Machine Learning to leverage data and identify complex interactions to address antimicrobial resistance in the ICU. ARTIFICIAL INTELLIGENCE IN THE ICU ICU Management & Practice 4 - 2023 145 pneumonia (Lopansri et al. 2019; Stevens et al. 2014). Parallel to the continuous search for the perfect discriminator of infection and inflammation, researchers have also turned to AI/ML to see if routinely collected healthcare data can be leveraged, with some interesting results. For example,routine colonisation cultures in the absence of infection are performed in many patients during their ICU stay and are often used when prescribing antimicrobials, the value of these cultures to guide antimicrobial therapy during a subsequent infection is subject of debate (Bredin et al. 2022; Barbier et al. 2016). Over the years, several risk factors have been identified that are linked to antimicrobial resistance (De Waele et al. 2018). Based on these risk models have been developed that try to discriminate bacterial from non-bacterial sepsis in children using eight routinely available parameters, which outperform currently available biomarkers (Lamping et al. 2018). Another study developed ML models to aid in the diagnosis of bacterial factors, clinical risk scores to predict the presence of AMR are being developed and tested but have not yet achieved widespread implementation (Burillo et al. 2019; Boyd et al. 2020). For this field as well, AI and routinely collected healthcare data are combined to fill a clinical need (MartínezAgüero et al. 2019; Pascual-Sánchez et al. 2020). Although promising, the results of these developed models have not yet been tested in clinical practice. Besides infection in COVID-19 patients, which are currently being tested in clinical practice (Rawson et al. 2021).
Once the right patient is identified, prescribing the correct antimicrobial entails assessing the possible causative pathogens and their potential resistance profile. While predicting infections with AMR organisms, expediting the identification of AMR organisms after obtaining a microbiological sample is another strategy to optimise antimicrobial prescribing. To this end, a variety of ML models combined with various phenotypic and genotypic resistance identification techniques for a range of sample types and organisms are being researched. Some models try to predict resistance based solely on a combination of limited microbiological (e.g., type of sample and gram stain) and demographic data (e.g., age and gender), while others are directly incorporated into the sample analysis flow of certain techniques such as liquid chromatography with tandem mass spectrometry or Raman spectroscopy (Feretzakis et al. 2020; Roux-Dalvai et al. 2019; Ho et al. 2019).
As demonstrated by these examples, it is clear that the interest in applying AI/ ML to the AMR domain is slowly growing. A recent review by Farhat et al. (2016) identified 676 AMR and AI/ML-specific publications, the majority (78.7%) being empirical papers and most of them being published after 2018, which is indicative of the juvenility of this nascent research domain. Despite the recency of this field, some research, such as incorporating AI/ML methods to augment and expedite current AMR diagnostic techniques, is envisioned to swiftly find its way into clinical practice once it has been thoroughly tested. Other research, such as predicting AMR from routinely collected healthcare data, might prove to be a bigger challenge to develop and implement.
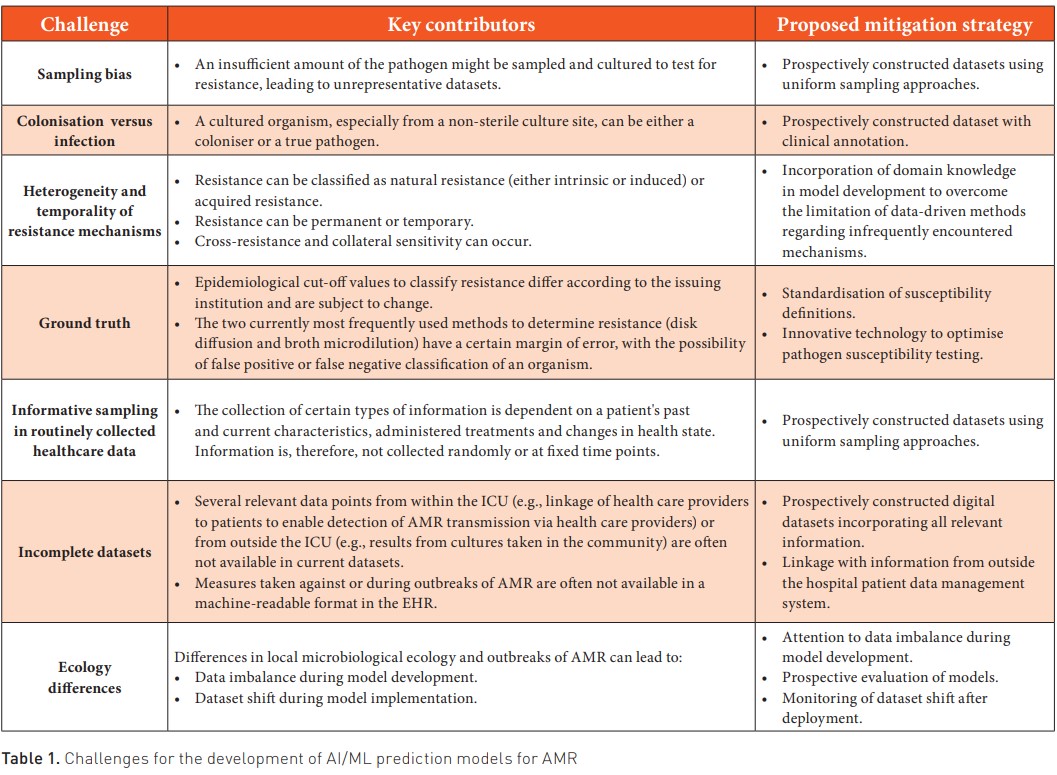
We identified seven main challenges for predicting AMR using AI/ML and routinely collected healthcare data. All seven are summarised in Table 1. For development, a first challenge is the potential sampling bias in routinely collected healthcare data. During clinical care, information is often not recorded on fixed time points nor randomly but only informatively, i.e., the collection of certain types of information is dependent on a patient’s past and current characteristics, administered treatments and changes in health state (Goldstein et al. 2016). In the case of AMR, for example, blood cultures will not routinely be drawn but only when there is a reason to do so. This might hinder the development of AMR prediction models using routinely collected healthcare data as they might not provide a proper AMR baseline status and warrants the development of prospectively designed use case-specific databases to avoid biases. Additionally, antimicrobial resistance is characterised by both intrinsic resistance (i.e., resistance that is universally expressed by a species to a certain antimicrobial, independent of previous antibiotic exposure, and not related to horizontal gene transfer) and induced resistance (i.e., resistance induced by antibiotic exposure or horizontal gene transfer) (Reygaert 2018). To complicate things even further, varying degrees of cross-resistance (when a single molecular mechanism induces resistance to multiple antimicrobial agents) or collateral sensitivity (when a single molecular mechanism induces susceptibility to multiple antimicrobial agents) can occur (Colclough et al. 2019). Adding to this complexity is the fact that AMR pathogens can spread across and within wards due to horizontal transmission, often with the caregivers as an intermediate. Purely data-driven ML models might not be able to identify these specific instances, as these do not occur frequently, and therefore, the necessary data to identify these relationships are often not present in routinely collected healthcare datasets. Alternative methods, such as hybrid AI or incorporating domain knowledge into ML, will have to be researched to incorporate these vital pieces of information into model development.
Considering the implementation of prediction models, antimicrobial resistance ecology might prove to be another challenge. As patient populations and patterns of antimicrobial resistance tend to vary between hospitals and between wards in the same hospital, the external validity of developed ML models will have to be carefully evaluated. But even when a model is used in the same ward as where it was developed, validity of the predictions should be carefully monitored, as outbreaks or horizontal transmission can quickly induce a change in local ecology, which in turn can lead to bias and thus malfunctioning of the prediction model due to a discrepancy between the population the model is used on and the population the model was developed on (Finlayson et al. 2021).
Finally, to be able to fully leverage AI/ML in the AMR domain, information will have to be included from outside the boundaries of the ICU. Antimicrobial treatments that were given by general practitioners and culture results from outside the hospital are just two examples of relevant information that can contribute to AMR research. Additionally, as several non-medical factors influence the occurrence of AMR, occupational and environmental information from other sources might also prove to be useful. Integrating all these different information sources will require data from these sources to be findable, accessible, interoperable and reusable (FAIR), as well as a more holistic approach to AMR from clinicians and researchers. Only then will we truly be able to approach antimicrobial resistance from a One Health perspective (Figure 1).
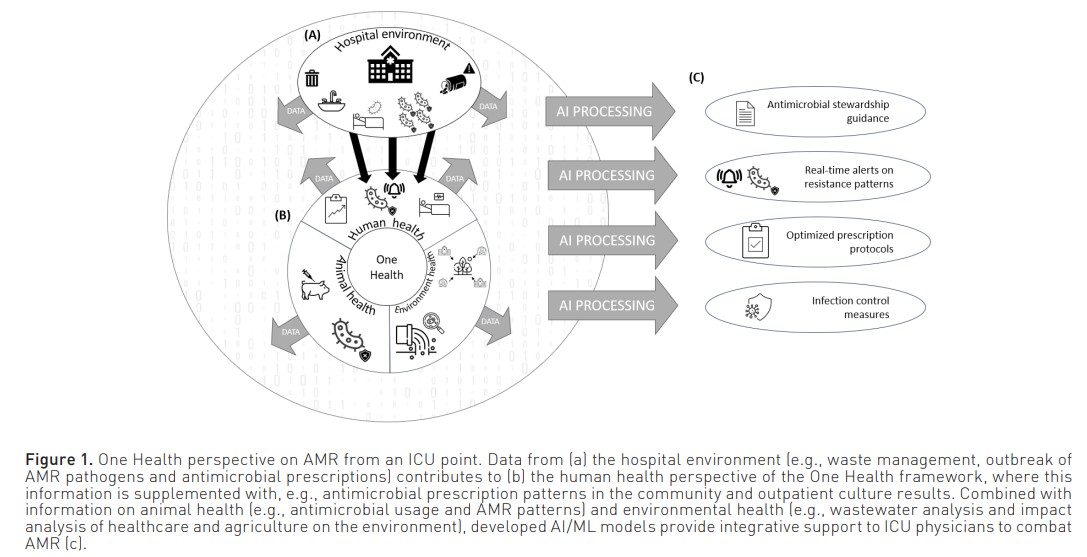
In summary, the possibility of AI and ML to leverage vast amount of data to identify complex interactions shows promise to address AMR in the ICU and support antimicrobial stewardship. Possible applications include discriminating inflammation from infection, optimising antimicrobial use, including empirical choices and dose selection, and predicting AMR pathogen involvement in acute infections. However, developing and implementing AMR prediction models in the ICU using a data-driven approach with routinely collected healthcare data poses several challenges due to, inter alia, informative data sampling and the intricacies of antimicrobial resistance. Other strategies, such as the development of goal-specific datasets and the inclusion of domain knowledge in model development, will need to be explored to overcome these issues. Ultimately, to enable a comprehensive AI-based approach to AMR from a "One Health" perspective, integration of diverse data sources beyond the ICU will be crucial.
Conflict of Interest
None.











