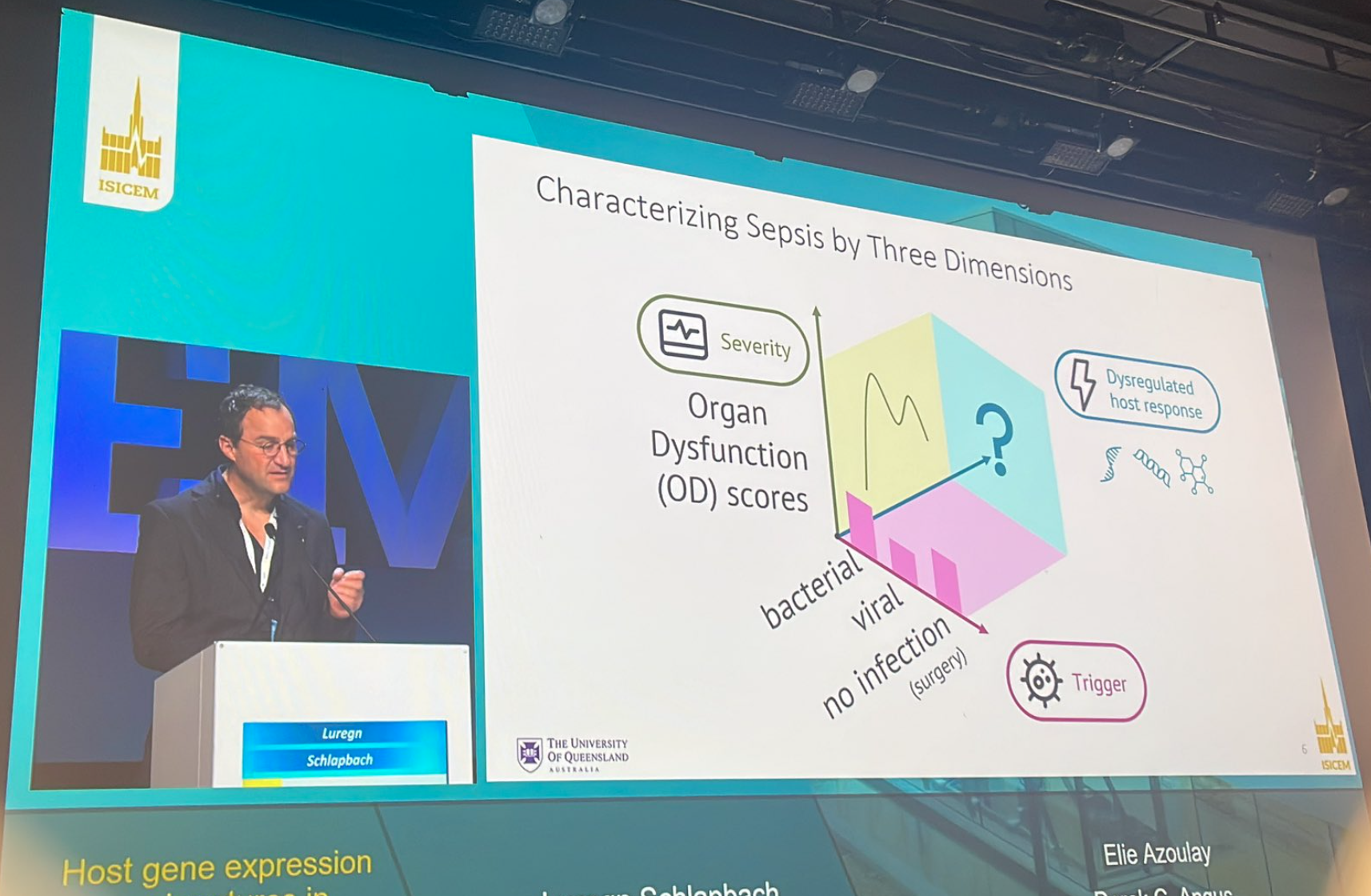
Findings presented at #ISICEM24 show that novel host transcriptomic signatures specific for bacterial and viral infection in children evaluated for sepsis can identify dysregulated host response leading to organ dysfunction.
The goal of the study was to create gene expression signatures in children with bacterial or viral infections to predict organ dysfunction, as current biomarkers for sepsis, defined as a severe response to infection, are insufficient.
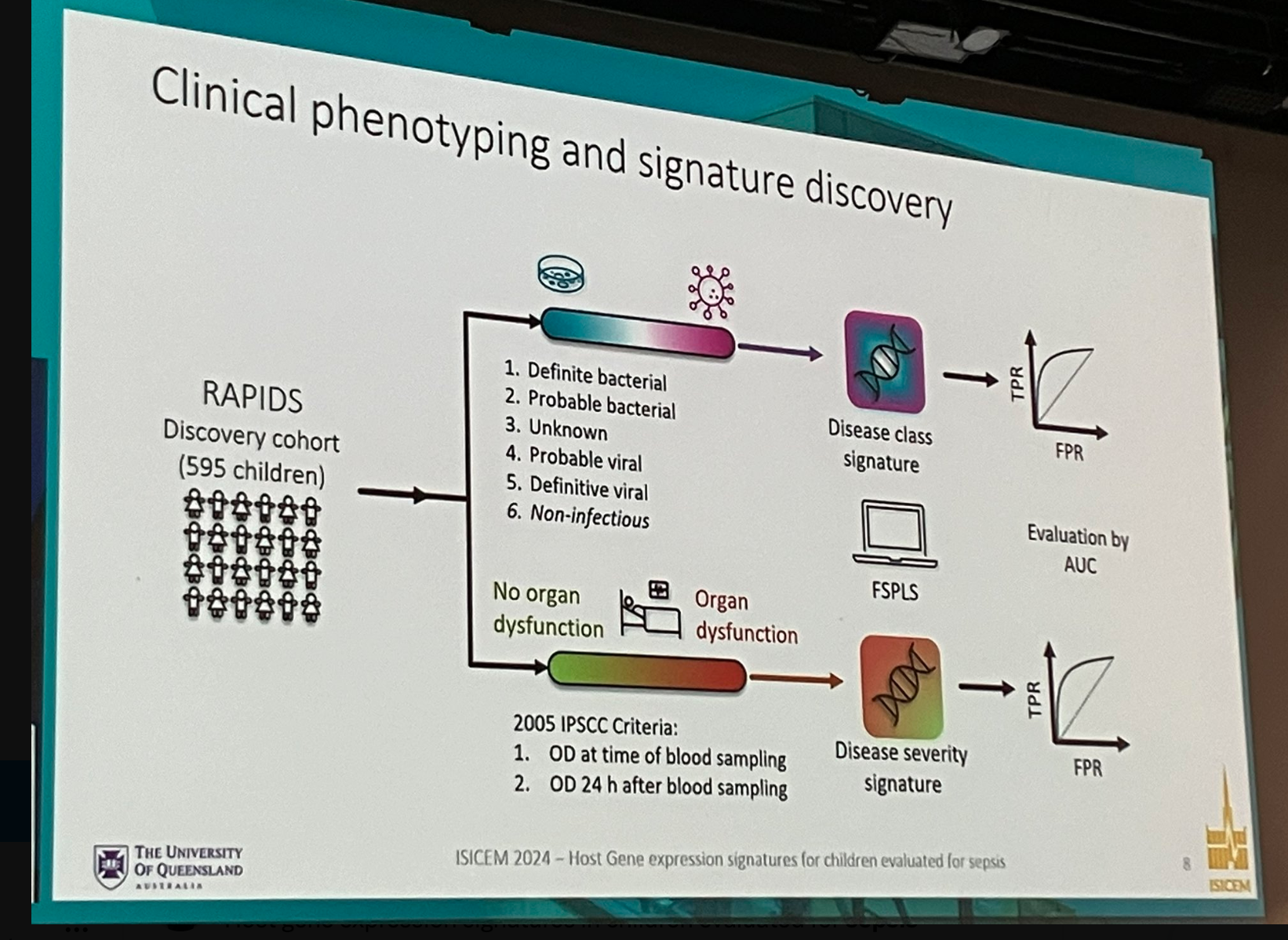
The study was conducted across emergency departments and ICUs in four hospitals in Queensland, Australia. Children one month to 17 years suspected of sepsis were recruited and underwent diagnostic testing, including blood cultures. Samples collected by March 31, 2020, formed the discovery cohort, while those collected afterward up to October 27, 2021, constituted the internal validation cohort named Rapid Paediatric Infection Diagnosis in Sepsis (RAPIDS). An external validation cohort was gathered from the European Childhood Life-threatening Infectious Disease Study (EUCLIDS).
Feature selection methods were applied to develop gene signatures for disease classification (bacterial vs viral infection) and severity (presence vs absence of organ dysfunction 24 hours post-sampling). The primary endpoint was the presence of organ dysfunction 24 hours after blood sampling in confirmed bacterial versus viral infections.
Nine hundred seven patients were enrolled in the study. Blood samples from 595 patients formed the discovery cohort, while samples from 312 children comprised the RAPIDS validation cohort.
A ten-gene signature for disease classification achieved an AUC of 94.1% in distinguishing between bacterial and viral infections in the RAPIDS validation cohort. Additionally, a ten-gene signature for disease severity achieved an AUC of 82.2% in predicting organ dysfunction within 24 hours of sampling in the RAPIDS validation cohort. When used together, these signatures predicted organ dysfunction within 24 hours of sampling with an AUC of 90.5% for patients with predicted bacterial infection and 94.7% for patients with predicted viral infection in the RAPIDS cohort. In the external EUCLIDS validation dataset (n=362), the combined signatures predicted organ dysfunction at the time of sampling with an AUC of 70.1% for patients with predicted bacterial infection and 69.6% (53.1–86.0) for patients with predicted viral infection.
In children being assessed for sepsis, newly developed host transcriptomic signatures tailored for bacterial and viral infections can effectively pinpoint the dysregulated host response associated with organ dysfunction.
Source: The Lancet, ISICEM
Image and Slides Credit: ISICEM
















