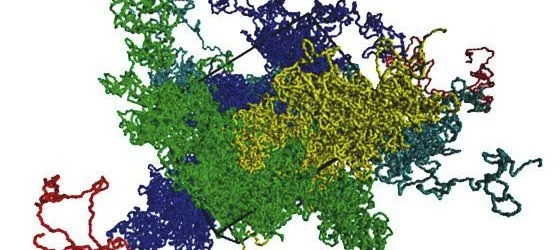
In the cell nucleus, DNA carries out its activities in a diluted state where it synthesises proteins and, despite resembling a messy tangle of thread, its structure is in actually governed by precise rules that are important for it to carry out its functions.
Customarily, biologists have studied DNA by observing it experimentally with a number of techniques. Only recently, these have been supplemented by research in silico, which is the study of DNA by means of computer simulations.
Though a new area of study, it has already contributed majorly to knowledge in this field. The state of the art of this novel but powerful approach has been assessed by Angelo Rosa, a theoretical physicist of the International School for Advanced Studies (SISSA) in Trieste, with the collaboration of Christophe Zimmer, an experimental physicist from the Pasteur Institute in Paris.
The systematic review of their findings has just been published in the journal 'International Review of Cell and Molecular Biology'.
Rosa believes this is first review of its kind, explaining that the review is mainly aimed at biologists as it makes minimal use of mathematical formulas that usually hamper reading and could also be of interest for physicists and mathematicians who are approaching this new field for the first time.
Reviewing 25 years of computational models the physicists observed that due to technical advances, the models had become increasingly sophisticated in this relatively short time. It was now possible to make far more detailed and predictive simulations, allowing science to lead the work of experimental researchers into previously unavailable directions.
"This is a useful tool which, without going into mathematical detail, provides the biologist with an overview of the type of studies that will increasingly complement the more traditional approaches" continues Rosa. "Today, for example, we already have software programmes which, starting from experimental data, allow us to reconstruct the structure of specific portions of chromosomes. I think that if computers continue to evolve as they have done until now -- and there's no reason to doubt this -- we'll be able to reconstruct entire chromosomes."
Rosa concludes that currently, the future prospects of in silico research into nuclear DNA are twofold. It will allow the detail understanding of the dynamics of gene expression (the details of protein synthesis), as well as the precise identification of where the chromosomes are when DNA unravels in the nucleus.
Source and image credit: Science Daily
6 January 2014
Latest Articles
Research, Genetics, cells, IT solutions, DNA, chromosome
In the cell nucleus, DNA carries out its activities in a diluted state where it synthesises proteins and, despite resembling a messy tangle of thread, its...