Magnetic Resonance Imaging (MRI) is one of the most effective tools used by doctors today to detect diseases and injuries. However, MRI scans can take hours or days to be interpreted and analysed and if there is a need for a detailed investigation, patients may be required to come back for a follow-up.
See Also: New Software for Fast and Accurate Interpretation in Radiology
To deal with this problem, researchers from the Texas Advanced Computing Center (TACC), The University of Texas Health Science Center (UTHSC) and Philips Healthcare have developed a new and automated platform that can return in-depth analysis of MRI scans within minutes. This can help minimise patient callbacks and can also result in significant savings. The new platform was demonstrated at the International Conference on Biomedical and Health Informatics in Orlando, Florida.
This new tool combines the imaging capabilities of the Philips MRI scanner with the processing power of the Stampede supercomputer. The Agave API Platform infrastructure has been used to facilitate communication, data transfer and job control between the two systmes.
"The Agave Platform brings the power of high-performance computing into the clinic," said William (Joe) Allen, a life science researcher for TACC and lead author on the paper. "This gives radiologists and other clinical staff the means to provide real-time quality control, precision medicine, and overall better care to the patient."
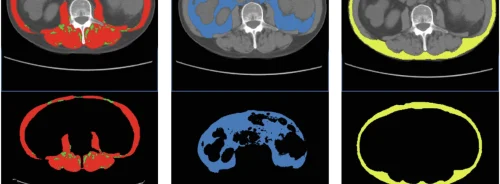
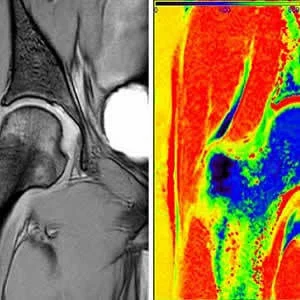
During the demonstration, the researchers performed MRI scans on a patient with a cartilage disorder. Data from the MRI passed through a server to Stampede where it went through the GRAPE ((GRAphical Pipelines Environment) analysis tool which characterises the scanned tissue and returns essential informatoin back to the clinician. The system uses a TI mapping process which converts data to imagery.
"Another potential of this technology is the extraction of quantitative, information-based texture analysis of MRI," aid Refaat Gabr, an assistant professor of Diagnostic and Interventional Imaging at UTHSC and the lead researcher on the project. "There are a few thousand textures that can be quantified on MRI. These textures can be combined using appropriate mathematical models for radiomics. Combining radiomics with genetic profiles, referred to as radiogenomics, has the potential to predict outcomes in a number diseases, including cancer, and is a cornerstone of precision medicine."
This new platform will enable doctors to capture a variety of biomedical data in real time and convert them into actionable insights for clinicians.
Source: University of Texas at Austin, Texas Advanced Computing Center
Image Credit: TACC, UTHRC, Philips Healthcare
Latest Articles
Magnetic Resonance Imaging, MRI scanner, Agave Platform, GRAPE
Researchers from the Texas Advanced Computing Center (TACC), The University of Texas Health Science Center (UTHSC) and Philips Healthcare have developed a new and automated platform that can return in-depth analysis of MRI scans within minutes