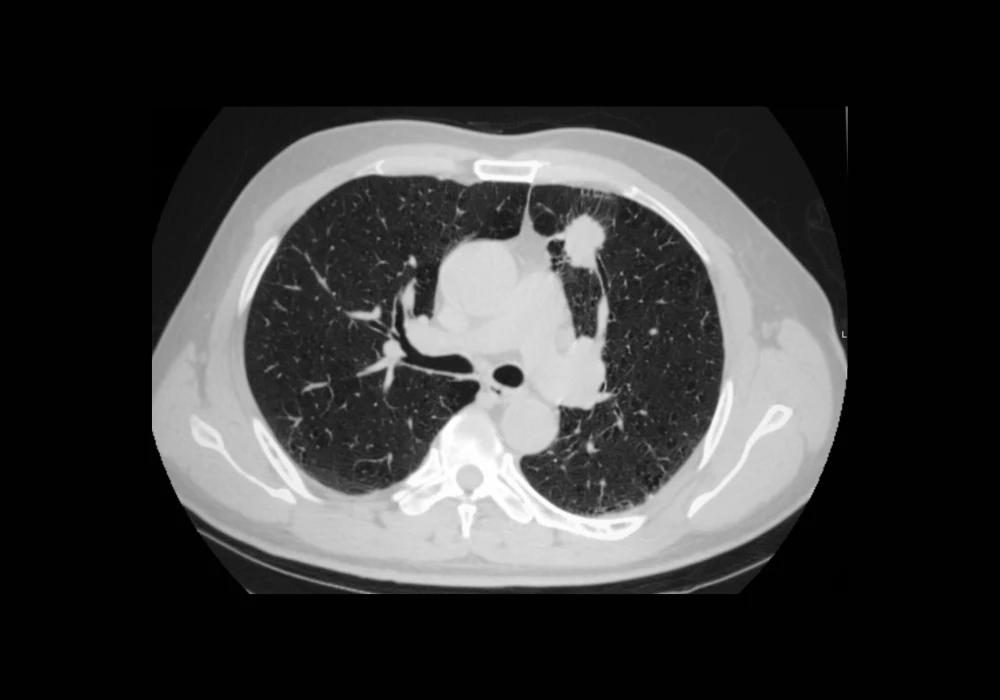
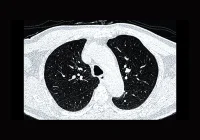
Accurate identification and quantification of lung cancer lesions is critical for diagnosis, staging, treatment planning and disease monitoring. While primary tumours receive most attention in radiological evaluations, patients often present with multiple lesions that must also be assessed for a complete picture of disease burden. Traditional manual segmentation remains laborious and subject to variability, making it less suitable for large-scale or real-world clinical use. A new automated pipeline now offers promising results for segmenting multiple lesions in computed tomography (CT) scans across diverse clinical settings, potentially improving consistency, sensitivity and efficiency in lung cancer management.
Capturing Clinical Complexity with Diverse Data
The automated pipeline was developed using 1,081 CT scans drawn from three distinct datasets encompassing both pretreatment and post-treatment cases. This included scans acquired with and without contrast agents, from 255 clinical sites and 8 different manufacturers, reflecting a wide range of protocols, scanner models and lesion presentations. Over 5,300 lesions were manually annotated by experienced imaging specialists following established radiological criteria. These included both primary tumours and metastases deemed visually discernible, clinically relevant and consistent with neoplastic morphology. The dataset was stratified for training and testing to reduce bias, with 868 cases used for model development and 213 for performance evaluation.
Must Read: Refining Nodule Management in Lung Cancer Screening
To support clinical relevance, only lesions measuring 10 mm or more were considered measurable, aligning with current RECIST 1.1 criteria. The model was also externally validated on an independent cohort of 188 real-world contrast-enhanced CT scans from the ChAImeleon European project. This validation cohort added further diversity in patient demographics, scanner types and acquisition parameters, strengthening the generalisability of the pipeline to routine clinical settings.
Architecture Tailored for Multi-Lesion Segmentation
The automated segmentation pipeline, known as LLSB_CFPR, was structured in three steps: thoracic region localisation, lesion segmentation and false positive (FP) reduction. First, the thoracic bounding box was extracted from each scan to reduce computational load and improve focus on the lungs, using lung masks generated by an open-source tool. Bounding box dimensions were adjusted to ensure lesions near pleural or mediastinal boundaries were not excluded.
The segmentation model was based on the nnU-Net architecture, selected for its high performance in medical image tasks. Modifications were made to accommodate multi-instance segmentation, including the generation of binary masks and the use of a 3D connected-components algorithm to distinguish individual lesions. The model was trained using five-fold cross-validation to ensure robustness and optimal parameter tuning.
To address the high false positive rate often seen in automated systems, especially in complex anatomical regions, a novel cascade false-positive reduction method (CFPR) was implemented. It combined two classifiers: one to exclude candidates located fully outside the lungs and another to validate candidates within the lung parenchyma. Both classifiers were tested with various image patch types, input configurations and network architectures, with the best performing versions selected through extensive experimentation.
Performance Across Internal and External Settings
On the internal test dataset, the pipeline achieved a lesion-level Dice similarity coefficient (DSC) of 81%, an image-level DSC of 76% and a lesion detection sensitivity of 85%. These results outperformed conventional architectures such as Swin-UNet, U-Net++ and UResNet when trained on the same data. Even with larger lesions, which are typically more clinically significant, the segmentation remained accurate, and volume measurements showed high agreement with manual annotations.
External validation on the ChAImeleon dataset confirmed the pipeline’s generalisability, with a lesion-level DSC of 78% and an image-level DSC of 73%. Lesion detection sensitivity remained high at 85%, although precision was slightly lower due to more variable imaging conditions. Despite this, the correlation between predicted and manual volume measurements remained strong, with minimal bias and narrow agreement limits.
Ablation studies further demonstrated the importance of each component. Introducing the thoracic bounding box alone improved image-level DSC by 5.4% and detection F1-score by 7.1%. Adding FP reduction through the dual-classifier CFPR approach led to a final 1% gain in DSC and a 3.2% gain in F1-score compared to the bounding box-only version. These incremental gains support the strategy of decomposing the FP filtering task into specialised components to target distinct error types.
The development of an automated, deep learning-based pipeline for multi-lesion lung cancer segmentation represents a significant step forward in radiological support tools. Its design accommodates the variability of real-world clinical data and addresses challenges often overlooked in controlled research environments. By enabling robust and consistent segmentation of multiple lesions across CT scans, the pipeline supports comprehensive disease assessment and has the potential to improve decision-making in oncology workflows. Future validation on larger, lesion-free cohorts and rare lesion subtypes will be essential to ensure readiness for widespread clinical adoption.
Source: European Radiology Experimental
Image Credit: iStock