Autosomal dominant polycystic kidney disease (ADPKD) is a hereditary condition that significantly impacts kidney function, affecting approximately 500,000 individuals in the United States and 12.4 million worldwide. This disease often involves multiple organs, including the liver, leading to polycystic liver disease (PLD). PLD is characterised by the presence of more than ten cysts in the liver and affects over 90% of ADPKD patients by their 50s. The severity of PLD can lead to debilitating symptoms for 2-5% of patients, sometimes necessitating liver cyst aspiration, fenestration, or even liver transplantation. As researchers and clinicians seek more precise methods for tracking ADPKD and PLD progression, the focus has shifted towards quantitative approaches using total kidney volume (TKV) and total liver volume (TLV) as prognostic biomarkers. However, these measurements face challenges in early-stage disease detection, prompting the need for better techniques to measure and characterise liver cysts directly. This article explores the advancements in automated liver cyst segmentation using deep learning, particularly the nn-UNet-based framework.
The Challenge of Manual Liver Cyst Segmentation
Manual marking and sizing of liver cysts in PLD are tedious, error-prone, and time-consuming for radiologists, especially when patients have numerous cysts. The process involves variability in image intensity due to factors like coil positioning, B1-inhomogeneities, and artefacts, making MRI analysis challenging. While CT scans offer more homogeneous imaging, they provide limited contrast compared to MRI T2-weighted imaging, making small cysts challenging to identify without contrast injection, which is not ideal for patients with chronic kidney disease. Various segmentation methods have been proposed, such as thresholding, region growing, and interactive manual segmentations. However, these methods have not demonstrated sufficient efficiency and accuracy for routine evaluations.
Adopting deep learning (DL) frameworks, particularly the nn-UNet-based approach, has shown promise in providing swift and precise liver cyst segmentation on MRI for PLD patients. This method utilises advanced algorithms trained on datasets from PLD patients undergoing biennial MRI monitoring and has been validated against expert observers, demonstrating high test-retest reproducibility.
Deep Learning Framework for Liver Cyst Segmentation
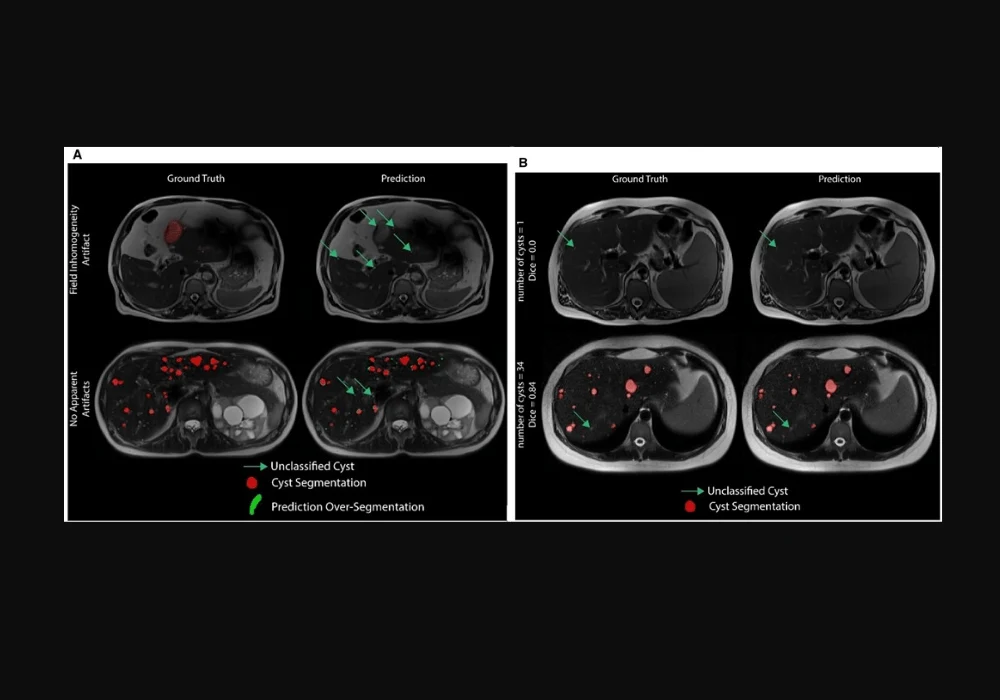
The nn-UNet framework is an encoder-decoder architecture with skip connections, instance normalisation, and leaky ReLU activation functions. It utilises a linear combination of Dice and cross-entropy loss functions to optimise segmentation performance. The training process involved annotating cysts in 40 ADPKD patients by experts using ITK-Snap, creating ground-truth segmentation masks for various MRI sequences. The model's performance was assessed on internal and external test sets, comparing its outputs to radiologist ground-truth masks using metrics like the patient-level Dice (PDice) and voxel-level true positive rate (VTPR).
The study demonstrated that the nn-UNet framework could significantly reduce the time required for expert observers to correct model outputs, enhancing the efficiency of liver cyst segmentation. The model achieved high agreement with manual segmentation and demonstrated reproducibility in biomarkers for disease analysis, such as cyst count and cyst fraction.
Enhancing Model Performance and Overcoming Limitations
Despite the promising results, the nn-UNet framework faced performance discrepancies across different MRI sequences. It performed best on T2-weighted sequences due to their excellent cyst-to-liver contrast but showed poorer performance on T1-weighted and SSFP sequences. To address this limitation, the study plans to implement a multi-channel model trained on multi-sequence input MR data and fine-tune the loss functions for better segmentation accuracy.
Another challenge was the variability in cyst detection, especially when there were few cysts. The study extended the evaluation metrics to patient-level and voxel-level assessments to provide a more robust evaluation of the algorithm's performance. Future work will focus on incorporating object-detection parameters, such as bounding boxes, to enhance the detection of individual cysts and improve overall segmentation accuracy.
The primary goal is to estimate biomarkers like cyst counts, volumes, and surface areas to assess disease severity and progression. The study demonstrated that the nn-UNet model could reliably reproduce biomarkers in early disease stages with fewer cysts. However, the model's performance degraded with an increased number of cysts, highlighting the need for larger and more diverse training datasets.
The nn-UNet-based deep learning framework offers a promising approach for efficient liver cyst segmentation in ADPKD patients. The model-assisted approach reduces the time required for radiologists to detect and segment cysts, showing high agreement with manual segmentation and excellent test-retest reproducibility. These advancements pave the way for more reliable and efficient quantitative biomarkers, facilitating the development of therapies aimed at slowing or arresting liver cyst growth in ADPKD patients. As training datasets continue to expand and model architectures are refined, the potential for early disease detection and improved patient outcomes will only increase, marking a significant step forward in the management of ADPKD and PLD.
Source & Image Credit: Radiology Advances