ICU Management & Practice, Volume 17 - Issue 1, 2017
The specialty of intensive care medicine grew out of the realisation that critically ill patients needed more attention and specialised treatment than could be provided on a general ward, and that many of these patients had similar clinical problems and processes, so management would be facilitated if they were grouped together in one place. Since those early days, intensive care medicine has grown rapidly with major advances in technology and understanding of disease pathogenesis and physiology. Progress in therapeutic interventions has, however, been less marked. One of the reasons behind the lack of effective new therapies relates to problems in performing randomised clinical trials in the very heterogeneous ICU patient populations. Indeed, since the birth of intensive care medicine, we have tended to group patients with similar signs and symptoms together under “umbrella” diagnoses, such as “sepsis”, “acute respiratory distress syndrome”, “acute renal failure”, ignoring the considerable heterogeneity within these groups in terms of individual characteristics, such as age, comorbid conditions, and genetic predisposition to disease; disease severity and degree of immune response; and individual variations in response to treatment. Performing randomised controlled trials in such mixed groups of patients will almost inevitably result in an inconclusive result as some patients in each group will respond to the therapy and others will not (Vincent 2016a).
Indeed we are increasingly aware that on the
ICU, as across all other medical fields, patients must be treated as
individuals and not as diseases. We have perhaps been too concerned with
defining syndromes and diseases and have somewhat “forgotten” the individual
people behind those conditions. We commonly hear phrases such as “he’s septic”,
“she’s a diabetic”, “where’s the ARDS patient?”, encouraging this attitude of
defining patients by their diagnoses, but we need to look behind the group
label and see the individual patient so that we can select the most appropriate
treatment for that person at that moment in time. This personalised approach to
medicine is not new; indeed, more than 2400 years ago, Hippocrates had already
noted the importance of individual characteristics in the development and
progress of disease and evaluated each patient and adjusted treatment according
to their “constitution, age, physique, the season of the year, and the fashion
of the disease” (Hippocrates, Nature of Man). Basic vital signs and variations in
physiological parameters, such as body temperature, heart rate and respiratory
rate, have also been used for centuries to assess a patient’s response to
therapy. As medicine has progressed, increasingly more complex parameters have
been used to predict outcome and adjust therapy, such as blood pressure and
cardiac output. In another attempt to help characterise patients, biomarkers
have been developed and studied as potential risk, diagnostic and prognostic
indicators for various conditions, including sepsis and acute kidney injury
(AKI) (McMahon and Koyner 2016; Pierrakos and Vincent 2010) although problems
of specificity and availability have limited their widespread use.
See Also: Precision Medicine in Sepsis
These relatively non-specific and simple methods
are now being complemented by more advanced techniques as, with the huge technological
advances of the last decade or so, we have begun to enter a whole new era of
personalised medicine. Genomic, transcriptomic, proteomic, and metabolomic profiling
techniques are enabling patients’ risks of disease and likely response to
treatment to be more closely identified, such that the treatment(s) most likely
to benefit that patient can be selected. For example, using genomic expression
profiling, Wong et al. (2015) identified two subgroups of children with septic
shock, one of which had increased mortality when prescribed corticosteroids. Similarly,
using whole genome amplification on blood samples from patients included in the
PROWESS study (Bernard et al. 2001), Man et al. (2013) identified two subgroups
of patients with different responses to treatment with drotrecogin alfa
(activated). The personalised medicine approach is now being applied to
clinical trials, helping select more specific groups of patients who are most
likely to respond to an intervention rather than the heterogeneous populations of
the past. For example, a study comparing granulocyte-macrophage
colony-stimulating factor (GM-CSF), an immunostimulating drug, with placebo, is
currently ongoing in patients with sepsis, but enrolling only patients
identified as being immunosuppressed based on their human leucocyte antigen
(HLA)-DR level (clinicaltrials.gov/ct2/show/NCT02361528) Such studies will
help, finally, to identify new therapies and interventions for conditions, such
as sepsis, in which multiple clinical trials in heterogeneous patients groups
have so far failed. Importantly, as these ‘omic techniques become more widely
used, costs will decrease. Drug development prices may also decrease as study
populations are more carefully defined, making trials more efficient.
Hand in hand with new analytic technology has come improved informatics capability, enabling sophisticated analysis of the large sets of patient data (demographic, physiological, laboratory and new ‘omic data) being collected, aided by national and international collaborations. Simulated models are also being developed to test suggested interventions on “virtual” patients or groups of patients, informing drug development and clinical trial design. The integration of all these data into “supermodels” (Brown 2015) may ultimately enable a physician to access a personalised treatment plan for every individual. These intelligent models will be able to update and adjust recommendations automatically as new data are received.
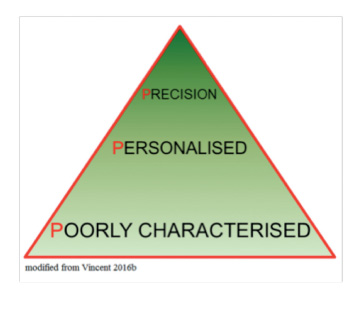
Clearly, this is still a somewhat futuristic view of personalised medicine in the ICU. Nevertheless, as we are increasingly able to better characterise patients, our ability to identify subgroups within subgroups will increase until we reach the point at which each subgroup consists of just one patient (Gattinoni et al. 2016). This will be true precision medicine, in which medical treatments will be customised to an individual’s molecular and genetic makeup. Although this approach is already being used in oncology, in the ICU environment, with the very rapid changes that occur in patient status, requiring regular treatment adjustment and thus necessitating repeated phenotypic profiling, true precision medicine is still some way off. Nevertheless, the progress from poorly characterised patient groups to personalised medicine is already a huge advance (Vincent 2016b).
References:
Bernard GR, Vincent JL, Laterre PF et al. (2001) Efficacy and safety of recombinant human activated protein C for severe sepsis. N Engl J Med, 344(10): 699-709.
Brown SA. (2015) Building SuperModels: emerging patient avatars for use in precision and systems medicine. Front Physiol, 6: 318.
Gattinoni L, Tonetti T, Quintel M (2016) Improved survival in critically ill patients: are large RCTs more useful than personalized medicine? We are not sure. Intensive Care Med, 42(11): 1781-3.
Man M, Close SL, Shaw AD et al. (2013) Beyond single-marker analyses: mining whole genome scans for insights into treatment responses in severe sepsis. Pharmacogenomics J, 13(3): 218-26.
McMahon BA, Koyner JL (2016) Risk stratification for acute kidney injury: Are biomarkers enough? Adv Chronic Kidney Dis, 23(3): 167-78.
Pierrakos C, Vincent JL (2010) Sepsis biomarkers: a review. Crit Care, 14(1): R15.
Vincent JL (2016a) Improved survival in critically ill patients: are large RCTs more useful than personalized medicine? No. Intensive Care Med, 42(11): 1778-80.
Vincent JL (2016b) Individual gene expression and personalised medicine in sepsis. Lancet Respir Med, 4(4): 242-3.
Wong HR, Cvijanovich NZ, Anas N et al. (2015) Developing a clinically feasible personalized medicine approach to pediatric septic shock. Am J Respir Crit Care Med, 191(3): 309-15.