Parkinson’s Disease (PD) is identified as the world’s fastest-growing neurodegenerative disorder, affecting approximately 10 million individuals globally. The urgency for effective disease-modifying strategies is underscored by significant gaps in understanding the early molecular events of PD and the absence of reliable biomarkers in easily accessible bio-fluids. One of the critical challenges in PD research is the lack of effective biomarkers that can reliably detect the disease in its early stages. Early detection is crucial for initiating interventions that can potentially slow or halt disease progression.
In a recent study, mass spectrometry identified 23 distinct proteins that significantly differentiated PD patients from healthy controls. These proteins are implicated in inflammatory pathways, suggesting early involvement in PD pathophysiology. Machine learning models applied to these proteins achieved remarkable accuracy (100%) in classifying PD and control samples, indicating strong potential for early detection.
Limitations in Current Understanding and Biomarker Development
Current research highlights significant gaps in understanding the molecular events that precede the clinical onset of PD. These gaps hinder the development of precise disease-modifying therapies and effective biomarkers. Traditional biomarker approaches, such as cerebrospinal fluid (CSF) analysis of proteins like SAA, show promise but are limited by variability in peripheral blood and the need for invasive sampling procedures.
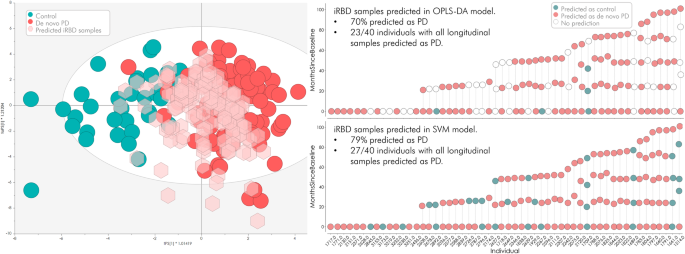
Current research highlights cerebrospinal fluid (CSF) SAA as a promising candidate for detecting neurodegenerative diseases. It exhibits high sensitivity (up to 74%) and specificity (up to 93%) in identifying prodromal stages like idiopathic REM sleep behaviour disorder (iRBD). However, its application is limited to CSF due to variability issues in peripheral blood.
Advantages of Mass Spectrometry in Biomarker Discovery
Mass spectrometry-based proteomics offers a more comprehensive approach to biomarker discovery compared to traditional antibody-based methods. It allows for the unbiased measurement of all expressed proteins in a sample, providing deeper insights into disease mechanisms. This technology enables researchers to identify biomarkers associated with PD pathology across different stages, from prodromal to clinical disease. To address these limitations, mass spectrometry-based proteomics emerges as a robust tool for identifying biomarkers. Unlike antibody-based multiplex technologies, which rely on selected panels and exhibit variability, mass spectrometry allows for unbiased measurement of all expressed proteins, providing deeper insights into disease mechanisms.
Validation and Longitudinal Assessment in Prodromal Stages
Further validation efforts included expanding the sample size and longitudinal assessment of iRBD subjects. The biomarker panel maintained a high prediction rate (79%) for identifying individuals who would later develop PD, even up to seven years before motor symptoms manifest. This longitudinal approach underscores the predictive power of the identified biomarkers.
The identified biomarkers provide insights into key pathophysiological mechanisms underlying PD. Elevated levels of Complement C3, for instance, correlate with motor dysfunction severity and cognitive decline. Downregulation of Wnt-signaling pathway proteins (such as DKK3 and PPP3CB) is associated with worsening motor symptoms. Increased stress-related proteins (e.g., HSPA5) indicate significant endoplasmic reticulum stress in early PD stages. Furthermore, independent upregulation of SERPIN family proteins (SERPING1, SERPINF2, SERPINA3) suggests heightened inflammatory activity, while decreased levels of progranulin (GRN) may indicate reduced neuroprotection.
Key findings from the study
- Elevated levels of Complement C3, correlating with motor dysfunction severity and cognitive decline.
- Downregulation of Wnt-signaling pathway proteins (DKK3 and PPP3CB), associated with higher motor symptom scores.
- Increased levels of stress-related proteins (such as HSPA5), indicating significant endoplasmic reticulum stress in early PD stages.
- Independent upregulation of SERPIN family proteins (SERPING1, SERPINF2, SERPINA3), suggesting heightened inflammatory activity.
- Decreased levels of progranulin (GRN), potentially indicating reduced neuroprotection.
The study’s integrated approach of multiplexed biomarker analysis and machine learning enhances the specificity and sensitivity of early PD detection. Future directions include validating these biomarkers in larger and independent cohorts, refining the biomarker panels for clinical utility, and advancing toward neuroprevention trials. These efforts aim to translate findings into actionable tools for early diagnosis and intervention in PD.
Source Credit: Nature Communications
Image Credit: iStock