Disease subtyping, crucial for personalised medicine, involves categorising diseases based on genetic, molecular, or clinical attributes. This enhances treatment efficacy and patient outcomes. The differentiation is vital for tailored treatments and understanding disease mechanisms across medical disciplines. While subtyping aids in effective treatment and discovering potential cures, it's particularly essential for diseases like diabetes mellitus and neurodegenerative disorders, where treatments vary significantly. However, some diseases like influenza don't require subtyping based on causal viruses.
Efforts to categorise diseases have historically relied on systems like the International Classification of Diseases (ICD). However, with increasing data, automated approaches are necessary to correct errors in large knowledge bases. Initiatives like the Open Targets (OT) platform integrate diverse datasets for disease subtyping. Existing methodologies, mainly rule-based, inherit disease levels from annotations and ontologies. A recent paper published in the Journal of Biomedical Informatics proposed a data-driven machine learning approach for OT to predict disease subtypes, offering scalable and interpretable solutions. This involves creating a target matrix, deriving predictive features, training a machine learning model, and iteratively validating predictions. The goal is to identify potential novel disease subtypes within existing databases, aiding future research and treatment strategies.
Utilising Open Targets Data for Automated Disease Subtyping and Prediction
Data from Open Targets, as of July 2022, was utilised for disease subtype prediction. Primary datasets included associationByOverallDirect, diseaseToPhenotype, associationByDatasourceDirect, and diseases. Disease subtypes were defined using the "has_children" attribute in the OT diseases dataset. The dataset underwent preprocessing, removing irrelevant diseases and aggregating similar ones. Features were extracted from OT direct evidence data sources, including genetic associations, phenotype counts, and literature evidence. Engineered features were also generated, such as aggregated statistics and evidence source ratios. Additionally, deep learning text features were derived using a pretrained biomedical language model.
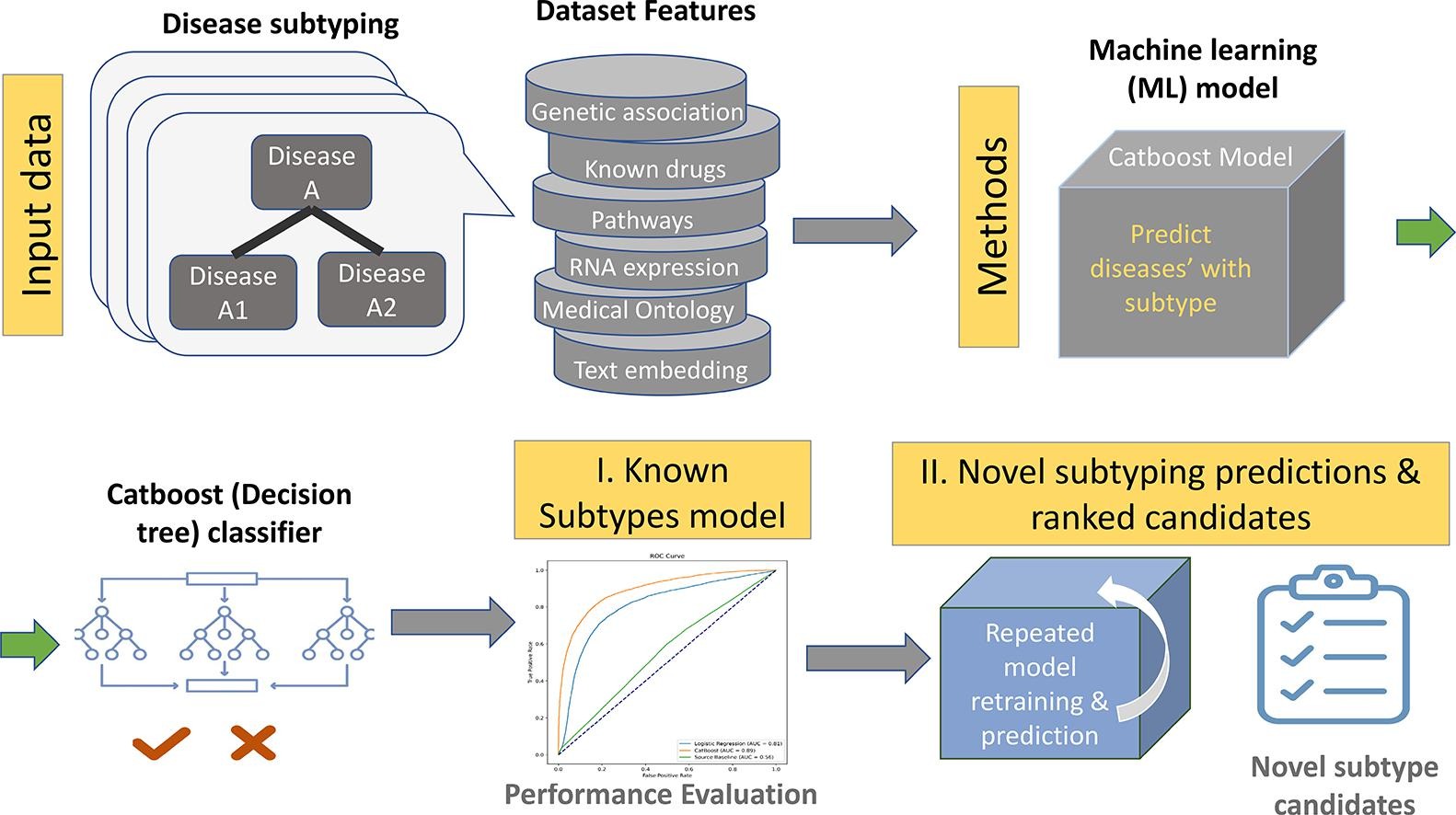
Models, including logistic regression, K-nearest neighbours, and tree models, were trained using scikit-learn and CatBoost. Stratified 5-fold cross-validation was employed to evaluate model performance, with features importance summarised using SHAP values. Deep learning text features were generated using BioLORD-STAMB2-v1, enhancing model performance. Overall disease population prevalence from the UK Biobank was considered but did not significantly impact model performance. The final model provided interpretable predictions of disease subtypes, aiding in medical research and treatment development.
Enhancing Predictive Performance and Overcoming Annotation Challenges
The authors’ hypothesis suggests that diseases with distinct subtypes can be identified based on intrinsic disease aspects and meta-features related to research. For instance, cancer diseases driven by somatic mutations differ from those influenced by genetic variants. Single-gene disorders may split into early and late onset diseases, affecting clinical approaches. Features are designed to capture disease characteristics and meta-science aspects, such as research limitations and phenotypic differences. This objective approach aims to improve predictive performance compared to biassed approaches reliant on existing literature and annotations. Disease rarity, quantified by population prevalence and orphan disease classification, is considered but found to have low impact on model predictions. Despite high-quality medical ontologies, challenges persist in annotating diseases with similar symptoms stemming from different causes, particularly with ambiguous pleiotropic diseases. Manual identification and validation of disease subtypes are labor-intensive and biassed towards diseases prevalent in developed countries. To address these limitations, a novel approach integrating OT direct evidence with machine learning is proposed. This method identifies potential disease subtypes, providing a ranked list for expert validation. The implications extend to clinical diagnostics and drug development, offering better diagnoses, personalised treatments, and targeted research.
Annotating disease subtypes is crucial for refining therapeutic strategies and enhancing patient outcomes. The study demonstrates the feasibility of automatically characterising known disease subtypes and identifies diseases likely to have uncharacterized subtypes, laying the groundwork for further research and expert confirmation.
Source & Image Credit: Journal of Biomedical Informatics
Title Image Credit: iStock