Alzheimer's disease is characterised by the buildup of amyloid-β plaques in the brain. Amyloid PET imaging is crucial for diagnosis and treatment planning. While visual interpretation is common, it has limitations. Quantitative analysis, despite being more accurate, is operator-dependent. As disease-modifying treatments emerge, there's a growing need for efficient PET interpretation tools. Deep learning shows promise in automating this process, but current models often lack generalizability and require complex preprocessing. This study published in Radiology aimed to develop a deep learning model for amyloid PET interpretation, assess its performance across different data sets and tracers, and compare it with human reads.
Utilising Publicly Available Datasets for Automated Assessment of Amyloid Positivity in PET Scans
The study conducted imaging research with approval from institutional review boards and participant consent. It utilized publicly available datasets from projects like the Alzheimer’s Disease Neuroimaging Initiative (ADNI), Open Access Series of Imaging Studies 3 (OASIS3), the Centiloid Project, the Anti-Amyloid Treatment in Asymptomatic Alzheimer’s Disease (A4) study, and the Study to Evaluate Amyloid in Blood and Imaging Related to Dementia (SEABIRD). These datasets included PET scans with various tracers like fluorine 18 florbetapir (FBP), carbon 11 Pittsburgh compound B (PiB), fluorine 18 florbetaben (FBB), fluorine 18 flutemetamol (FMT), and fluorine 18-flutafuranol (previously 18F-NAV4694).
PET scans were preprocessed, and amyloid positivity was determined using established cutoffs. Additionally, manual reads were conducted for comparison, involving a radiology and nuclear medicine expert. The scans were labelled as amyloid positive or negative based on their mean cortical SUV ratio.
A convolutional neural network (CNN) named AmyloidPETNet was developed to assess the probability of amyloid positivity in PET scans. This CNN architecture was inspired by previous studies and consisted of five context modules with residual connections. These modules were designed to process input volumes of any size, making the model versatile for different PET scan formats. The CNN was trained on the labelled PET scans to learn patterns indicative of amyloid positivity.
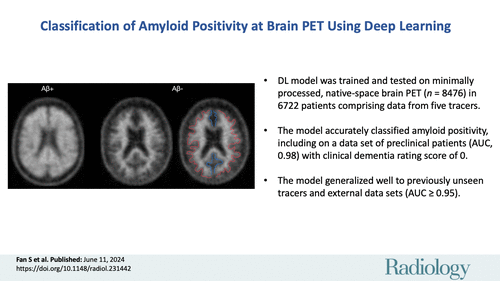
Image Credit: Radiology
Performance Evaluation of AmyloidPETNet for Predicting Amyloid Positivity in PET Scans
The performance evaluation of AmyloidPETNet involved several steps and analyses. First, the ADNI dataset was divided into training, validation, and test sets, ensuring balanced distributions of mean cortical SUV ratio and sex using anticlustering. The model underwent training for 100 epochs with an early stopping mechanism to prevent overfitting. Hyperparameter tuning was conducted, and the best-performing model was retrained five times to assess robustness. Additionally, the model was extensively validated on independent hold-out sets, including scans with unseen tracers, from various datasets like OASIS3 and A4.
The model's ability to predict amyloid positivity was evaluated across different scenarios. This included within the same tracer and dataset (ADNI), across different tracers within the same dataset (ADNI), across independent datasets using the same tracer as in the training dataset, and on data from different study datasets acquired with a different tracer than in the training set. Furthermore, performance evaluation based on patients' biological sex, Clinical Dementia Rating score, and age groups was conducted.
Comprehensive Statistical Analysis and Interpretability Assessment
For statistical analysis, various performance metrics such as accuracy, balanced accuracy, positive predictive value, negative predictive value, sensitivity, specificity, positive and negative likelihood ratios, F1 score, and area under the receiver operating characteristic curve (AUC) were calculated. Interreader agreement between AmyloidPETNet predictions and visual reads was assessed using Cohen's κ statistic. Bootstrap 95% confidence intervals were computed for all metrics, and statistical tests including the McNemar test, the generalised score statistic, and the DeLong test were employed for comparisons across different datasets. Visualisation and interpretability were addressed using Local Interpretable Model-agnostic Explanations to visualise model attention and ensure that the model was learning to classify based on relevant features. Finally, computational costs were estimated by measuring the average running time of the model on scans with and without graphics processing units to evaluate efficiency.
Robust Performance Across Diverse PET Tracers and Datasets for Amyloid Positivity Prediction
The study included 8476 PET scans from 6722 patients, with a median age of 70.9 years. Among these scans, 33.1% were amyloid-positive, and 85.3% of the patients were cognitively unimpaired. For model training and validation, 1743 scans from 861 patients were used, while 184 scans from 95 patients were used for internal testing. The external testing set comprised 6549 PET scans from 5769 patients.
AmyloidPETNet exhibited high accuracy and AUC when predicting amyloid positivity using the same PET tracer (18F-FBP) in the ADNI dataset, achieving an accuracy of 96.8% and an AUC of 1.0 in the training set. The model's performance remained robust on the validation and internal test sets, achieving AUCs of 0.97 on both datasets.
When predicting amyloid positivity using the same PET tracer across independent datasets (OASIS3 and the A4 study), AmyloidPETNet achieved AUCs of 0.95 and 0.98, respectively. Despite the relatively low amyloid-positive prevalence in these datasets, the model achieved high F1 scores and balanced accuracy.
Furthermore, AmyloidPETNet demonstrated promising performance when predicting amyloid positivity using different PET tracers across independent datasets. Despite being trained only on 18F-FBP PET scans, the model achieved high AUCs on 11C-PiB scans from OASIS3 and SEABIRD. Additionally, the model showed potential for 18F-FMT and 18F-NAV4694 scans from the Centiloid Project, although further experiments with larger samples are warranted to enhance performance assessments for these radionuclides.
Performance in Classifying Amyloid Positivity Compared to Expert Visual Reads
The model demonstrated robust performance across various scenarios. Most classification errors were within 10% of the amyloid-positive cutoff, with misclassified scans often exhibiting substantial cortical atrophy or poor image quality. Additionally, model performance remained consistent across different participant demographics, including sex, age groups, and Clinical Dementia Rating scores, with no statistically significant differences observed.
When comparing model classifications with expert visual reads, high agreement was observed in the Centiloid Project dataset, achieving an overall Cohen κ of 0.93. Only four disagreements were observed out of 127 scans, with three of these cases involving low confidence from the radiologist. In the A4 study, which contained more equivocal cases, model-physician agreement was lower (Cohen κ of 0.39). However, the model outperformed the radiologist when considering mean cortical SUV ratio, achieving a higher Cohen κ of 0.67.
Despite the challenges in equivocal cases, the model showed promising performance, particularly in accurately classifying cases near the cutoff point. Overall, the model demonstrated good agreement with expert annotations across different tracer datasets.
AmyloidPETNet: A Versatile and Accurate Deep Learning Model for Amyloid Positivity Prediction in PET Scans
One of the main advantages of AmyloidPETNet is its ability to provide rapid and accurate predictions of amyloid status regardless of imaging equipment, reconstruction algorithms, or Aβ tracer used. This could significantly impact both clinical and research settings, aiding in the diagnosis and treatment of patients with mild cognitive impairment, increasing diagnosis confidence, and assisting in the screening of interventions without the need for structural MRI or extensive computational resources.
Additionally, AmyloidPETNet demonstrated robust performance across different Aβ tracers, overcoming the challenges posed by the diversity of interpretation guidelines for each tracer. The model's attention maps revealed a diffuse focus on the gray-white matter boundary, reflecting typical patterns observed in amyloid PET scans.
Compared to previous studies, AmyloidPETNet showcased several improvements, including testing on large external datasets, generalization across various tracers, and the ability to process three-dimensional volumes of any size. The model's performance was comparable to human expert readers and could potentially enhance clinical decision-making in diagnosing Alzheimer's disease.
However, the study had limitations, including the exclusion of real-world clinical data with non-AD dementia and various comorbidities, as well as the lack of quantitative measures of Aβ pathology or prediction uncertainty. Nonetheless, AmyloidPETNet represents a significant advancement in automating the classification of amyloid PET scans, offering a reliable and efficient tool for nuclear physicians in clinical and research settings.
Source: RSNA Radiology
Title Image: iStock