Prostate cancer remains one of the most prevalent malignancies affecting men worldwide and poses a major burden on health systems. While biparametric MRI is recommended as a primary diagnostic approach, its standard assessment method—PI-RADS—has limitations, particularly a high false-positive rate. These limitations often result in unnecessary biopsies, with implications for patient safety and resource utilisation. To address this, a multimodal machine learning model was developed and validated across multiple centres to improve the prediction of clinically significant prostate cancer using imaging, clinical and radiomic features.
A Multimodal Approach to Diagnosis
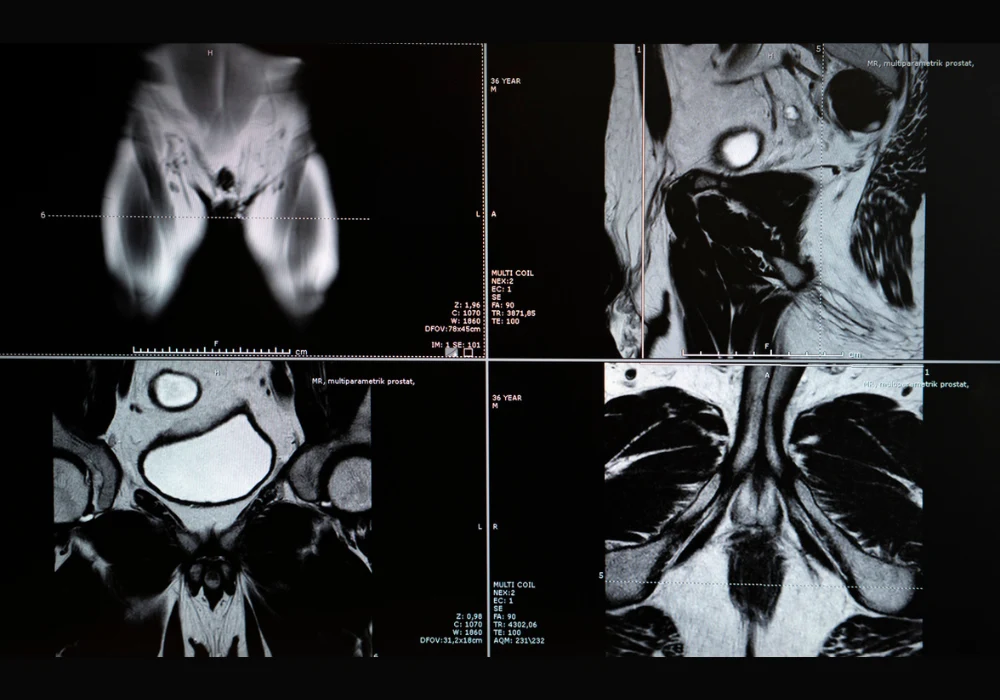
The multimodal model was trained using retrospective data from twelve clinical centres, incorporating T2-weighted, diffusion-weighted imaging and apparent diffusion coefficient sequences. It combined radiomic features, which quantify shape and texture, with clinical variables such as age, PSA levels, lesion location and PI-RADS scores. Radiomic extraction covered the entire prostate gland using segmentation masks generated by a deep learning model. Performance was assessed through retrospective and prospective datasets, ensuring temporal generalisability and robustness across scanner vendors and field strengths.
Must Read: Prostate Cancer Detection with Minimal MRI Data
The model consistently outperformed PI-RADS in detecting clinically significant cases, defined as ISUP grade 2 or higher. Compared with PI-RADS, the model achieved a higher AUC and specificity in both retrospective and prospective cohorts. At the same sensitivity level, the model reduced unnecessary biopsies by more than 20%, and at the same specificity, it detected more cases of clinically significant cancer. SHAP analysis confirmed that radiomic features, alongside PI-RADS and lesion location, significantly contributed to prediction performance.
Importance of Diversity and Generalisability
Sensitivity analysis demonstrated that performance depended heavily on the diversity of training data. Models trained on single-vendor datasets showed reduced generalisability, underscoring the importance of multicentric training. Scanner vendor and the presence or absence of an endorectal coil were considered in model variants, confirming the added value of multimodal training. Sequence-specific performance highlighted the superiority of biparametric MRI combinations over individual sequences alone.
Radiomics-only models underperformed compared to multimodal models that included clinical and semantic features. The learning curve analysis showed that model performance plateaued after using 50% of the available training data, indicating that future optimisation may not require full datasets. Despite this, limitations such as poor segmentation performance due to wide field-of-view diffusion sequences in some centres affected sensitivity. Consistent performance was maintained across age groups and scanner types, but local data quality affected outcomes in specific countries.
Clinical Impact and Future Directions
The model demonstrated strong calibration and decision curve performance, supporting its potential clinical utility. Compared to PSA density thresholds, it showed significantly higher sensitivity and specificity. As it builds upon rather than replaces PI-RADS scoring, and provides explainability via SHAP values, the model is well-suited for integration into current radiology workflows.
The approach avoids the need for lesion annotations and considers the whole prostate gland, which is especially relevant for multifocal prostate cancers. The work also contributes to the emerging literature that supports the use of noninvasive radiomics in clinical decision-making. Although further validation is needed in settings with variable radiologist expertise, the model’s robustness across vendors and imaging protocols makes it a promising tool for reducing unnecessary biopsies.
A multimodal machine learning model integrating clinical, semantic and radiomic features has shown substantial improvements in detecting clinically significant prostate cancer compared to standard PI-RADS scoring. By reducing false positives and increasing diagnostic accuracy, the model offers a valuable tool to support radiologists and improve patient outcomes. Further studies incorporating additional clinical variables could enhance its predictive power and support broader implementation in prostate cancer diagnostics.
Source: Radiology: Imaging Cancer
Image Credit: iStock