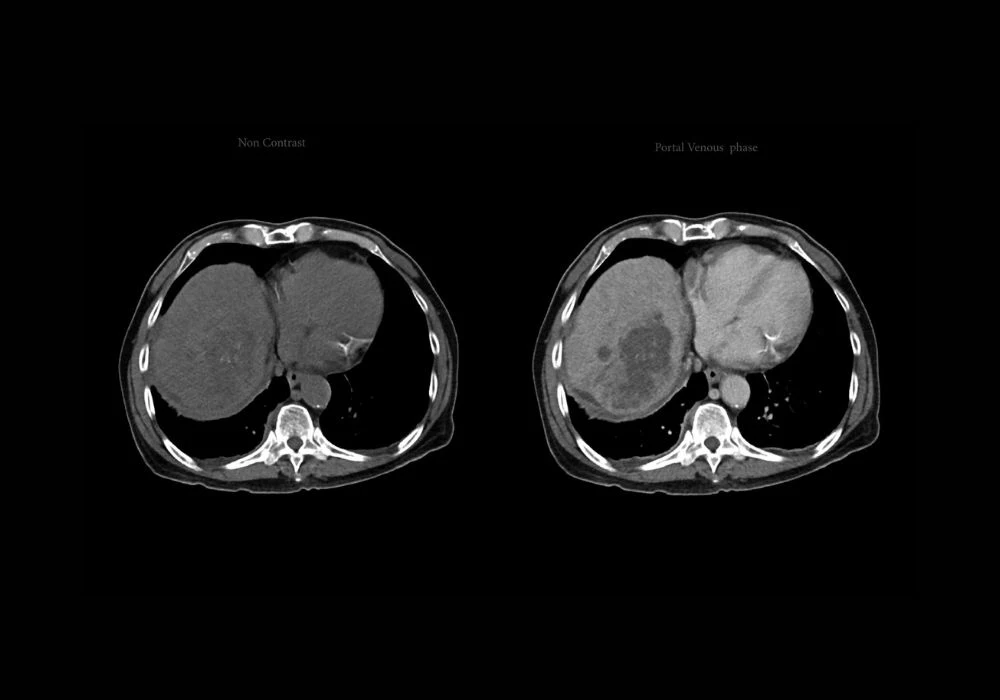
Hepatocellular carcinoma (HCC) remains a major global health challenge, being the third leading cause of cancer-related mortality. Its aggressive variants, particularly the proliferative subtype, are associated with poor prognoses and high recurrence rates post-surgery. Traditional diagnostic approaches rely heavily on invasive methods like histopathology, which come with inherent risks. While dynamic contrast-enhanced MRI (DCE-MRI) offers a noninvasive alternative, its full diagnostic potential is underutilised due to the subjective nature of image interpretation. To address these challenges, a novel deep learning (DL) model integrating self-supervised learning (SSL) with dynamic radiomics has been developed. This model enhances the precision and consistency of identifying proliferative HCC subtypes from multi-sequence DCE-MRI, offering new avenues for personalised treatment planning.
Enhancing Radiological Insight through Self-Supervised Deep Learning
Dynamic radiomics enables the extraction of time-resolved imaging features from DCE-MRI, offering deeper insights into tumour behaviour. However, conventional radiomics relies on manual annotation of regions of interest, which is both labour-intensive and prone to variability. To overcome this limitation, the pHCC-SSL model integrates a self-supervised approach that allows autonomous learning from raw imaging data without manual labelling. At its core, the model employs a sample-weighted full-convolution mask autoencoder (SW-FCMAE) to extract deep features, minimising redundancy and enhancing feature diversity.
The architecture follows a two-stage process. Initially, the model undergoes SSL pre-training to develop a foundational understanding of imaging characteristics. This is followed by a task-specific fine-tuning phase, where deep dynamic features are constructed by aggregating temporal data across multiple MRI sequences. These features are then processed using a self-attention mechanism, enabling the model to learn patterns indicative of tumour proliferation over time. This dynamic learning capability significantly enhances the model’s diagnostic accuracy.
Model Performance and Validation Across Diverse Clinical Settings
The pHCC-SSL model was trained and validated on data from 381 patients across two medical centres, split into training, internal testing and external testing sets. The results demonstrated robust performance, with the model achieving an AUC of 0.91 in the internal test set and 0.94 in the external test set. This outperformed models that did not utilise SSL, highlighting the benefit of the pre-training phase. Comparative assessments against single-sequence prediction models further confirmed the superior accuracy of the multi-sequence pHCC-SSL model.
Must Read: Advancing HCC Diagnosis with Combined Imaging
In addition to quantitative metrics, the model’s interpretability was enhanced using class activation mapping (CAM), which visually identified tumour regions most influential in predictions. The heat maps revealed focused activity within tumour zones, particularly in sequences that captured key vascular characteristics, supporting the model’s reliability in clinical evaluation.
Furthermore, statistical analyses demonstrated that the pHCC-SSL score was a more effective predictor of proliferative HCC than conventional markers like alpha-fetoprotein (AFP), which had significantly lower AUC scores. These findings underscore the added diagnostic value of integrating radiomics with advanced DL frameworks.
Clinical Implications and Future Considerations
The implementation of pHCC-SSL represents a significant advancement in noninvasive oncology diagnostics. By automating the extraction and interpretation of complex imaging data, the model reduces reliance on subjective assessments and manual inputs, thereby streamlining clinical workflows. Its ability to consistently identify proliferative HCC subtypes enables earlier intervention and better-informed therapeutic decisions, ultimately contributing to improved patient outcomes.
However, the study also recognises limitations. The retrospective nature introduces potential selection bias, and the prevalence of hepatitis B virus in the study population may limit generalisability to regions with different aetiological profiles. Additionally, while the model demonstrated high accuracy, its perfect performance on training data suggests possible overfitting. Expanding the dataset, incorporating multi-centre and multi-ethnic data and refining training strategies such as data augmentation and dropout techniques are essential next steps to enhance the model’s robustness and applicability.
The pHCC-SSL model signifies a promising shift towards AI-enhanced diagnostics in hepatocellular carcinoma. By leveraging self-supervised learning and dynamic radiomics, it offers a powerful tool for the precise and noninvasive identification of proliferative HCC. Its successful validation across multiple centres and its superior performance compared to traditional models reinforce its potential for clinical adoption. Continued research and broader validation will be key to fully realising its impact on personalised cancer care.
Source: Insights into Imaging
Image Credit: iStock