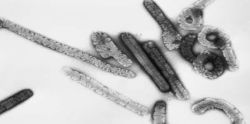
Researchers have developed a new approach to the diagnosis of tuberculosis (TB) that relies on direct sequencing of DNA extracted from sputum to detect and characterise the bacteria that cause TB. The new method eliminates the need for culturing bacteria in the laboratory, a time-consuming process.
The new technique is referred to as metagenomics, defined as the study of genetic material collected directly from environmental samples, the report said. The technique was developed by researchers working in the UK and Gambia. Results of their study have been published in the peer-reviewed journal PeerJ.
Metagenomics involves the use of genomic techniques to study microbial communities in their natural environments without the need to cultivate them in the lab. In this case, researchers used sputum samples taken from patients suspected of having TB. They detected sequences from the TB bacteria in all sputum samples they examined. The researchers also were able to assign the bacteria to a known lineage in seven of the samples.
Rapid Identification of TB Strain Key to Effective Treatment
Two sputum samples were found to contain sequences from Mycobacterium africanum, a species of Mycobacterium most commonly found in West African countries. Great diversity exists among species of Mycobacterium, the researchers said, noting that rapid identification of the precise pathogenic strain is critical for effective treatment of tuberculosis.
Conventional laboratory diagnostic methods for identifying TB usually takes weeks or months. In contrast, metagenomics would take only a few hours to do the work, according to Professor Mark Pallen, director of the research. He said that relying on lab culture "means using techniques that date back to the 1880s!"
Metagenomics combines the latest high-throughput sequencing technologies and some smart bioinformatics. "(This) allows us to detect and characterise the bacteria that cause TB in a matter of a day or two, without having to grow the bacteria, while also giving us key insights into their genome sequences and the lineages that they belong to,” Prof. Pallen explained.
Development of New Diagnostic Tools for Detecting TB
Prof. Pallen and his team believe that this new technique can help in the development of new diagnostic tools for detecting TB. In an earlier work, the team used metagenomics to successfully isolate pathogenic strains from human samples. For example, they successfully identified the genome of E.Coli, which caused an infectious outbreak last year.
In addition, metagenomics was used on historical samples when the research team isolated the genome of Brucella melitensis, a bacterium which causes an infection called brucellosis in livestock and humans, from a 700-year-old skeleton from Sardinia, Italy.
Pallen et al. said that more improvements were needed to fully integrate metagenomics in the diagnosis and analysis of infectious pathogens, although they noted that successful identification of the TB lineages "provides proof of its effectiveness".
Source: MedicalDaily.com
Image Credit: University of Southern California
The new technique is referred to as metagenomics, defined as the study of genetic material collected directly from environmental samples, the report said. The technique was developed by researchers working in the UK and Gambia. Results of their study have been published in the peer-reviewed journal PeerJ.
Metagenomics involves the use of genomic techniques to study microbial communities in their natural environments without the need to cultivate them in the lab. In this case, researchers used sputum samples taken from patients suspected of having TB. They detected sequences from the TB bacteria in all sputum samples they examined. The researchers also were able to assign the bacteria to a known lineage in seven of the samples.
Rapid Identification of TB Strain Key to Effective Treatment
Two sputum samples were found to contain sequences from Mycobacterium africanum, a species of Mycobacterium most commonly found in West African countries. Great diversity exists among species of Mycobacterium, the researchers said, noting that rapid identification of the precise pathogenic strain is critical for effective treatment of tuberculosis.
Conventional laboratory diagnostic methods for identifying TB usually takes weeks or months. In contrast, metagenomics would take only a few hours to do the work, according to Professor Mark Pallen, director of the research. He said that relying on lab culture "means using techniques that date back to the 1880s!"
Metagenomics combines the latest high-throughput sequencing technologies and some smart bioinformatics. "(This) allows us to detect and characterise the bacteria that cause TB in a matter of a day or two, without having to grow the bacteria, while also giving us key insights into their genome sequences and the lineages that they belong to,” Prof. Pallen explained.
Development of New Diagnostic Tools for Detecting TB
Prof. Pallen and his team believe that this new technique can help in the development of new diagnostic tools for detecting TB. In an earlier work, the team used metagenomics to successfully isolate pathogenic strains from human samples. For example, they successfully identified the genome of E.Coli, which caused an infectious outbreak last year.
In addition, metagenomics was used on historical samples when the research team isolated the genome of Brucella melitensis, a bacterium which causes an infection called brucellosis in livestock and humans, from a 700-year-old skeleton from Sardinia, Italy.
Pallen et al. said that more improvements were needed to fully integrate metagenomics in the diagnosis and analysis of infectious pathogens, although they noted that successful identification of the TB lineages "provides proof of its effectiveness".
Source: MedicalDaily.com
Image Credit: University of Southern California
Latest Articles
TB, bacteria, tuberculosis, metagenomics, pathogens
Researchers have developed a new approach to the diagnosis of tuberculosis (TB) that relies on direct sequencing of DNA extracted from sputum to detect and...